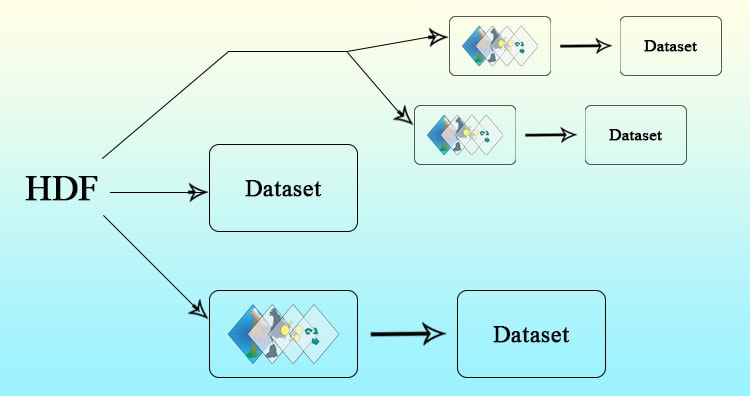
Hierarchical Data Format HDF multidimensional data, which can grow in space and time dimension. It is specially designed by National Center for Supercomputing Applications (NCSA) for storing scientific data in mosaic datasets. You may also look over GRIB, netcdf and all multidimensional data.
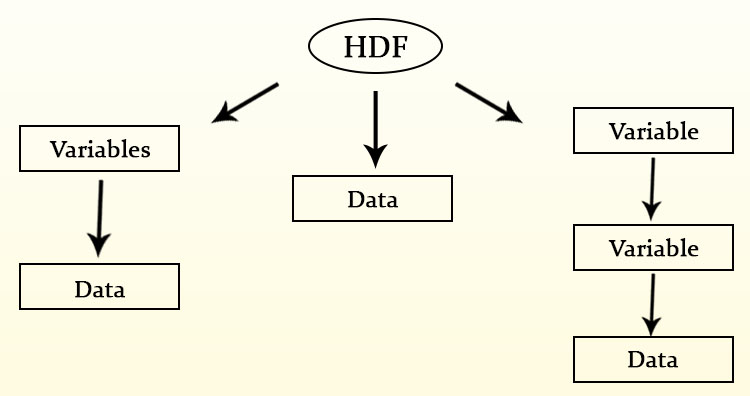
Here the data is stored in hierarchical format i.e. in tree like structure. It contains band information in variables. Variables may have one or more variable. Each band data in this variables may differ in size and dimension. Also each data can contain information of different regions.
HDF 4 (Hierarchical data format release 4) – HDF MultiDimensional Data
HDF supports two formats, both are completely different and NOT compatible. It supports multidimensional arrays, raster images and table data. It has some limitations as it supports multiple (arrays, table and images) data hence makes complex API.
HDF 5 (Hierarchical data format release 5)- HDF MultiDimensional Data
This format is designed to overcome the limitations of HDF 4. This structure consist of datasets and groups.
- Groups, which are container structures which can hold datasets
- Datasets, which are multidimensional arrays of a homogeneous type.
Get information of HDF multidimensional data bands-
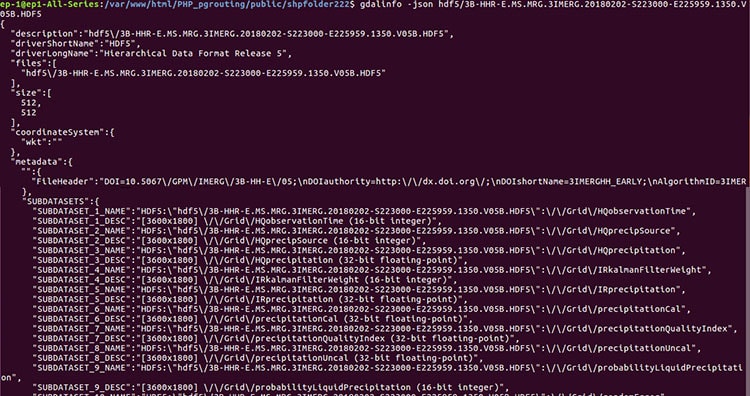
As data is arranged in hierarchical format, we need to run two command to get band details, both from GDAL utility.
1. Gdalinfo- This commands gives the information about number of subdatasets present in file. Each sub-dataset detail is stored in SUBDATASET_n_NAME metadata item. The related description is stored in SUBDATASET_n_DESC metadata item.
2. Gdal_translate- The gdal_translate utility is used to import the bands or convert in desired format. Here we have converted HDF data in NetCDF format.

Hope this article help you to understand HDF data format. You can also visit multidimensional data for more information. Please let us know if you require any help, by commenting in given comment box.